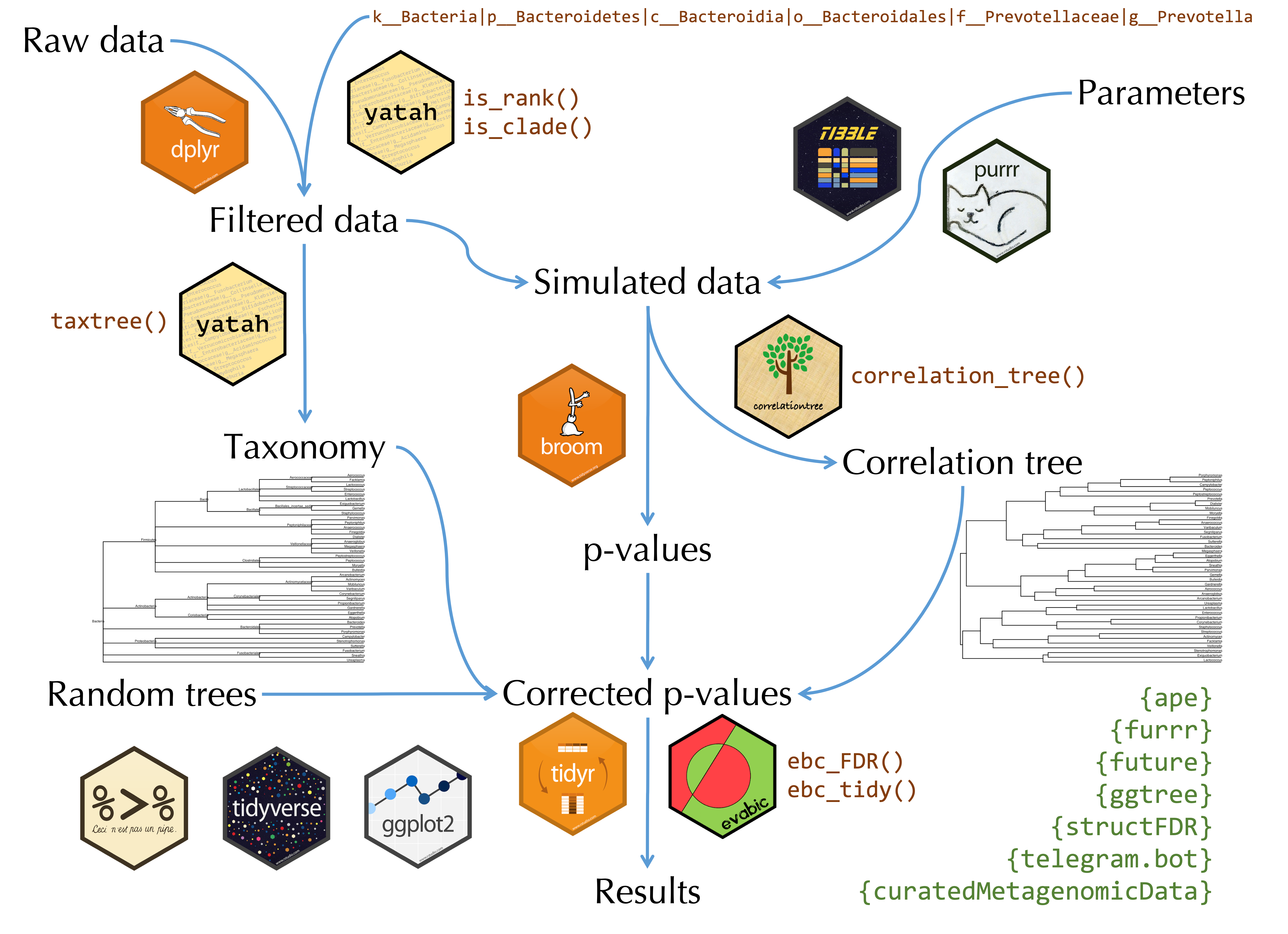
Workflow
| Phylum | Class | Order | Family | Genus | S001 | S002 | S003 | S004 | S005 | … |
|---|---|---|---|---|---|---|---|---|---|---|
| Actinobacteria | Coriobacteriia | Coriobacteriales | Atopobiaceae | Atopobium | 84 | 0 | 12 | 54 | 0 | … |
| Actinobacteria | Coriobacteriia | Eggerthellales | Eggerthellaceae | Eggerthella | 2 | 0 | 0 | 7 | 0 | … |
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | 525 | 7 | 134 | 753 | 0 | … |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | 88 | 1770 | 1490 | 119 | 2136 | … |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 0 | 0 | 138 | 4 | 0 | … |
| Firmicutes | Negativicutes | Veillonellales | Veillonellaceae | Dialister | 152 | 4 | 2 | 192 | 0 | … |
| Firmicutes | Negativicutes | Veillonellales | Veillonellaceae | Megasphaera | 402 | 0 | 4 | 102 | 0 | … |
| Fusobacteria | Fusobacteriia | Fusobacteriales | Leptotrichiaceae | Sneathia | 302 | 0 | 35 | 272 | 0 | … |

The z-scores \(\mathbf{z} = \Phi^{-1}(\mathbf{p})\) are smoothed using the following hierarchical model:
\[\mathbf{z} | \mathbb{\mu} \sim \mathcal{N}_n \left( \mathbb{\mu}, \sigma^2 \mathbf{I}_m \right) \qquad \mathbf{\mu} \sim \mathcal{N}_m\left(\gamma \mathbf{1} , \tau^2 \mathbf{C}_{\rho} \right)\]
where \(\mathbf{C}_{\rho} = \left(\text{exp} (−2\rho \mathbf{D}_{i,j} )\right)\) with \(\mathbf{D}\) the patristic distance matrix between taxa from the tree. By applying Bayes’s formula:
\[\mathbf{z} \sim \mathcal{N}_m \left(\gamma \mathbf{1},\tau^2 \mathbf{C}_{\rho} + \sigma^2 \mathbf{I}_m\right)\]
\[\mathbb{\mu}^* = \left(\mathbf{I}_m + \frac{\sigma_0^2}{\tau_0^2} \mathbf{C}_{\rho_0}^{-1}\right)^{-1}\left(\frac{\sigma_0^2}{\tau_0^2} \mathbf{C}_{\rho_0}^{-1}\gamma_0 \mathbf{1} + \mathbf{z}\right)\] Finally, a permutation-based FDR control is applied on \(\mathbb{\mu}^*\)
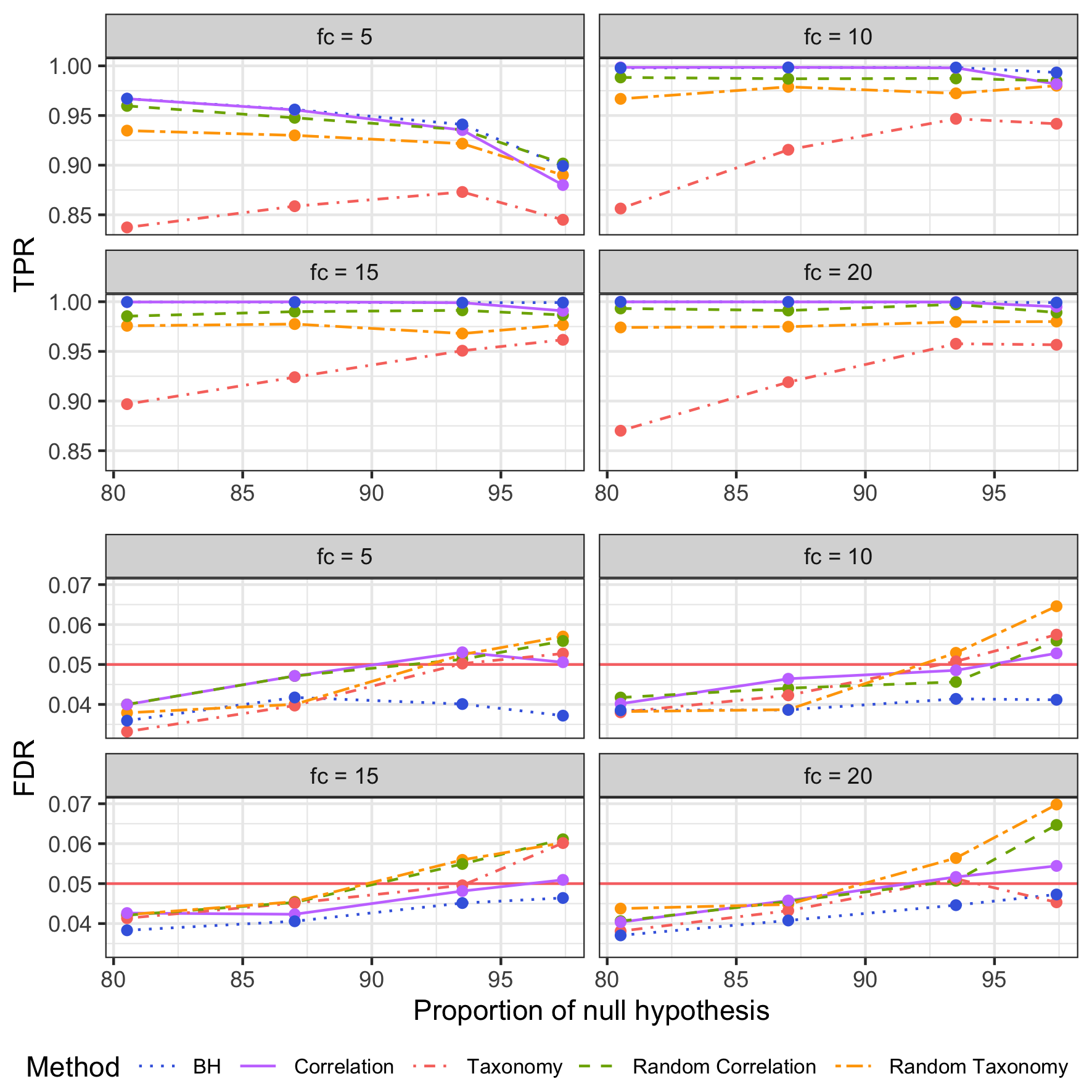
The tree choice has little impact on detection power
Benjamini-Hochberg procedure is still the most powerful method and the only one which respects the FDR control
The ease of creating R packages greatly increases the reproducibility of analysis
tidyverse and especially list-columns allow to write elegant and efficient R code when manipulating non-standard structures (trees, statistical model outputs…)
Poster made with pagedown
📄 Xiao, Jian, Hongyuan Cao, and Jun Chen. False discovery rate control incorporating phylogenetic tree increases detection power in microbiome-wide multiple testing. Bioinformatics 33.18 (2017): 2873-2881.
📄 Bokulich, Nicholas A., et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Science translational medicine 8.343 (2016): 343ra82-343ra82.
📄 Opstelten, Jorrit L., et al. Gut microbial diversity is reduced in smokers with Crohn’s disease. Inflammatory bowel diseases 22.9 (2016): 2070-2077.